Arrival of the Fittest: Solving Evolution's Greatest Puzzle (18 page)
Read Arrival of the Fittest: Solving Evolution's Greatest Puzzle Online
Authors: Andreas Wagner

It’s not enough to know, of course, that a library contains a virtually inexhaustible supply of books describing solutions to a particular problem. We also need to find out where these solutions are and how they are organized—in meticulous stacks or thrown together in unruly piles. And for that we need to move beyond laboratory experiments. Even though such experiments can create and test impressive numbers of different proteins, these numbers vanish into insignificance compared with those found in nature, which churns out new proteins every day in countless trillions of live organisms. Every one of these organisms harbors thousands of proteins, and each is only the last link in an unbroken chain of protein creation that goes back billions of years.
Protein scientists have been aware of this bounty for many years. And they throw themselves at it with gusto, like kids in a warehouse-sized candy store. And what these scientists have learned about protein creation in thousands of different organisms goes far beyond laboratory experiment. The oxygen-ferrying hemoglobin we already encountered in the bar-headed goose illustrates how far.
The humble function of hemoglobin, binding and releasing oxygen as it shuttles between the lung and the body’s tissues, belies its near-universal importance. Hemoglobin is a member of a large family of oxygen-binding proteins, the
globins,
which are vital not only to us but also to many other mammals, birds, reptiles, and fish.
36
Countless generations have elapsed, parents followed by their children, grandchildren, and innumerable generations of great-grandchildren, since all these organisms shared a common ancestor. During this succession of generations, the DNA that encodes hemoglobin and all other proteins has replicated countless times. The copying errors it suffers each generation are rare—about one for every forty million copied DNA letters in our cells
37
—but given enough time, all genes in a genome will suffer errors that alter the proteins they encode.
An altered amino acid text that prevents a globin from folding also prevents oxygen from traveling where it is needed. In other words, it spells death. But an altered protein does not always lose its function and meaning. Some alterations impair neither folding nor function, and get passed on to the next generation.
38
Over thousands and millions of generations, copy error after tolerable copy error can thus accumulate and slowly change a protein’s amino acid sequence.
FIGURE 11.
Proteins change in time
Figure 11 shows a snippet of ten amino acids from human hemoglobin and from three of our animal relatives.
39
Each letter in the figure is taken from the twenty-letter alphabet that scientists use to abbreviate amino acids, V for valine, A for alanine, and so on. We and our closest relatives, the chimpanzees, shared a common ancestor some five million years, or roughly 200,000 human generations, ago—a huge amount of time compared to a human life span, but precious little in evolution.
40
And because little time means few errors, the globin text of chimpanzees has not changed much since then. In the figure’s snippet, the only difference is that chimp globins harbor the amino acid glutamate (letter E, in black) in place of the human alanine (A).
The human lineage split from the mouse lineage some eighty million years ago. Mouse globins thus had more time to accumulate changes than chimps’, which shows in the two amino acid differences of figure 11 between mice and humans. The chicken lineage separated from us even longer ago—almost three hundred million years—and accrued six altered amino acids.
41
Millions of other organisms harbor globins, not only warm-blooded vertebrates but reptiles, frogs, fish, sea stars, mollusks, flies, worms, and even plants. Some of these organisms grow on the same twig of life’s gigantic tree and have a recent common ancestor. Their globin texts shared most of their journey through time, split only recently, and are still similar. Others lie on different branches, share more distant ancestors, and harbor globins with different texts.
42
But however different these texts are, each of them works just fine, since otherwise it would not have survived. Each surviving text encodes a different solution to the problem of binding oxygen.
43
And every millennium that life continues, it travels further and further through the library of proteins, discovering ever-new globin texts in its blindly groping evolutionary journey.
44
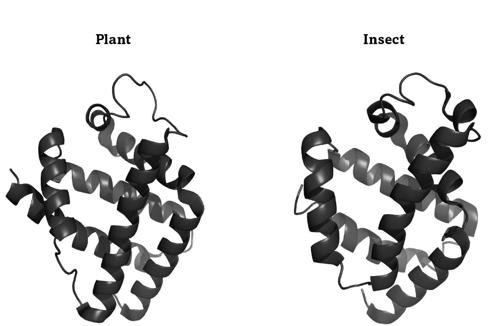
FIGURE 12.
Two hemoglobins with similar folds
To see how far this journey has already led the globins, consider some of our most distant relatives: plants, some of which indeed have globins, even though they have no blood.
45
Legumes like soybeans, peas, and alfalfa can extract vital nitrogen from its nearly unlimited supply in the air. (Most other plants need to extract nitrogen from the soil, where it is often scarce, unless a farmer has applied fertilizer.) For this purpose, legumes employ bacteria that live in clumps of tissues around their roots, and that harbor a special enzyme that converts airborne nitrogen gas into ammonium, the same ammonium that nitrogen fertilizers contain. This ingenious symbiosis has only one problem: Atmospheric oxygen destroys the enzyme. To protect those enzymes, plants manufacture globins, which keep oxygen safely away from the bacteria.
Plants and animals dwell on different major branches of life’s tree, because their common ancestor lived more than a billion years ago. Their globins are staggeringly different, which reflects their long and separate evolutionary journey. For instance, the globins from lupins and insects differ in almost 90 percent of their amino acids. Yet these globins not only bind oxygen, they also still fold into the very similar shapes of figure 12. The fold on the left is from a legume, the one on the right from a midge, a tiny two-winged fly. Both proteins have several spiral-staircase-like helices that are arranged similarly, such as the two helices that run in parallel from the upper left to the lower right. The image does not do full justice to just how similar these globins are. If you rotated these molecules to place one exactly above the other, their atoms would occupy almost exactly the same places. Despite more than a billion years of separation, these globins still fold the same way.
The amino acid differences between these globins are extreme, but not unusual. Even globins from some animals, for example those of clams and whales, can differ in more than 80 percent of their amino acids.
46
Despite these differences, though, these and thousands more globins from other organisms are connected by a network of unbroken paths through the protein library, paths that began at their common ancestor, took one amino-acid-changing step at a time, but left the text’s meaning unchanged. You will recognize a theme that we already encountered in the metabolic library, where evolution could travel far and wide without losing the meaning of a metabolic phenotype. The steps evolution takes through the protein library are different—single amino acid changes instead of horizontal gene transfer—but the principle is the same. A
genotype network
connects globins and extends its tendrils far through the protein library. Evolution can explore the library along this network without falling into the deadly quicksand of molecular nonsense.
When it comes to forming vast and far-reaching genotype networks, globins are not an exception but the rule. Enzymes with the same fold, catalyzing the same reaction, and sharing the same ancestor
typically
share less than 20 percent of their amino acids. We know this because scientists have mapped the location of texts encoding thousands of known enzymes in the library. By cataloging these texts, we can map the paths of genotype networks in the library, which reveals that some networks can reach even further through the library than globins, and none more so than that of TIM barrel proteins. Their name is an acronym for triose phosphate isomerase, an enzyme that helps extract energy from glucose, and their fold is called a barrel because its sheets and helices are arranged like the staves of a barrel. The stunning fact is that some enzymes with this fold do not have a single amino acid in common. They occupy opposite corners of the protein library—texts that do not share a single letter—yet carry the same chemical message.
47
Proteins like these are a bit like innumerable versions of
Hamlet,
all of them equally stageable, while sharing only a few hundred—or even none—of the play’s four thousand lines.
Thousands of proteins from nature’s laboratory tell a similar same story: When a problem can be solved with new proteins, be they enzymes, regulators, or transporters like hemoglobin, the number of solutions is too large to count. And all of those proteins are connected by a vast network of amino acid texts that spread throughout the protein library. We know thousands of proteins from some of these networks, but they are grains of sand on a vast beach of the unknown—most of the many trillions of proteins that share the same phenotype. Some of these unknown proteins belong to long-extinct organisms. But most have not even formed yet. The four billion years of life were not nearly enough time—they would suffice to create only some 10
50
proteins, a vanishingly small fraction of all texts in the protein library.
48
Life’s enormous tree and all its proteins, however vast and beautiful, is but a smeared reflection in a filthy mirror, a faint shadow of the vast Platonic realm that genotype networks inhabit.
In chapter 3, we saw that genotype networks help evolution’s billions of readers explore different and far-flung neighborhoods of the metabolic library. Through these networks, some of the library’s explorers find innovative texts with new phenotypes, even though others get knocked off a network and die. Genotype networks might do the same for proteins, but only if the neighborhoods of the protein library are diverse.
49
Otherwise, an evolving population of proteins might as well stay wherever it is. No need to explore the library if its different stacks host the same books.
Do the shelves near each protein in the library contain texts with similar meaning, a bit like modern suburbs with their identical cookie-cutter homes? Or is each neighborhood more like a medieval village, with unique buildings and their individual charm, containing proteins with unique new functions? Until recently, we had no idea, even though decades of protein research allow us to answer this question with computers that can mine mountains of protein data.